Amphisphaeriales Hawksw. & O.E. Erikss., Syst. Ascom. 5(1): 177 (1986)
MycoBank number: MB 90458; Index Fungorum number: IF 90458; Facesoffungi number: FoF 00672;
Amphisphaeriales has been accepted in consecutive studies using multigene phylogenies in the subclass Xylariomycetidae (Senanayake et al. 2015, Samarakoon et al. 2016b, Hongsanan et al. 2017, Crous et al. 2018b). However, based on previous studies, Wijayawardene et al. (2018a) accepted 11 families in Amphisphaeriales, i.e. Amphisphaeriaceae, Apiosporaceae, Beltraniaceae, Clypeophysalosporaceae, Hyponectriaceae, Melogrammataceae, Oxydothidaceae, Phlogicylindriaceae, Pseudomassariaceae, Sporocadaceae and Vialaeaceae, while Cainiaceae, Coniocessiaceae, and Iodosphaeriaceae were accepted in Xylariomycetidae families incertae sedis. In this study, Amphisphaeriales is a moderately supported clade sister to Xylariales and comprises 17 families viz. Amphisphaeriaceae, Apiosporaceae, Beltraniaceae, Castanediellaceae, Clypeophysalosporaceae, Cylindriaceae, Hyponectriaceae, Iodosphaeriaceae, Melogrammataceae, Oxydothidaceae, Phlogicylindriaceae, Pseudomassariaceae, Pseudosporidesmiaceae, Pseudotruncatellaceae, Sporocadaceae, Vialaeaceae, and Xyladictyochaetaceae. The divergence time for Amphisphaeriales has been estimated as 133 MYA. This number did not include Vialaeaceae because this family goes to Xylariales in MCC tree (Fig. 2). Currently there are 17 families and 88 genera in this order (this paper).
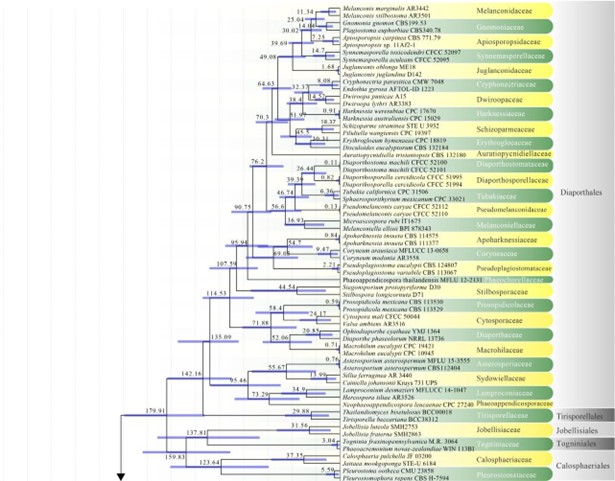
Figure 2 – The maximum clade credibility (MCC) tree, using the same dataset from Fig. 1. This analysis was performed in BEAST v1.10.2. The crown age of Sordariomycetes was set with Normal distribution, mean = 250, SD = 30, with 97.5% of CI = 308.8 MYA, and crown age of Dothideomycetes with Normal distribution mean = 360, SD = 20, with 97.5% of CI = 399 MYA. The substitution models were selected based on jModeltest2.1.1; GTR+I+G for LSU, rpb2 and SSU, and TrN+I+G for tef1 (the model TrN is not available in BEAUti 1.10.2, thus we used TN93). Lognormal distribution of rates was used during the analyses with uncorrelated relaxed clock model. The Yule process tree prior was used to model the speciation of nodes in the topology with a randomly generated starting tree. The analyses were performed for 100 million generations, with sampling parameters every 10000 generations. The effective sample sizes were checked in Tracer v.1.6 and the acceptable values are higher than 200. The first 20% representing the burn-in phase were discarded and the remaining trees were combined in LogCombiner 1.10.2., summarized data and estimated in TreeAnnotator 1.10.2. Bars correspond to the 95% highest posterior density (HPD) intervals. The scale axis shows divergence times as millions of years ago (MYA).
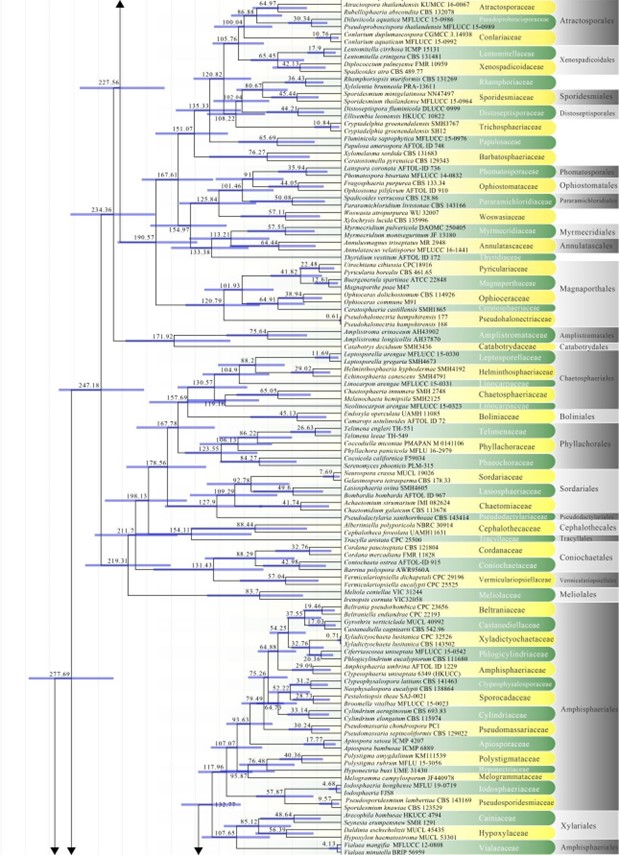
Figure 2 – Continued.
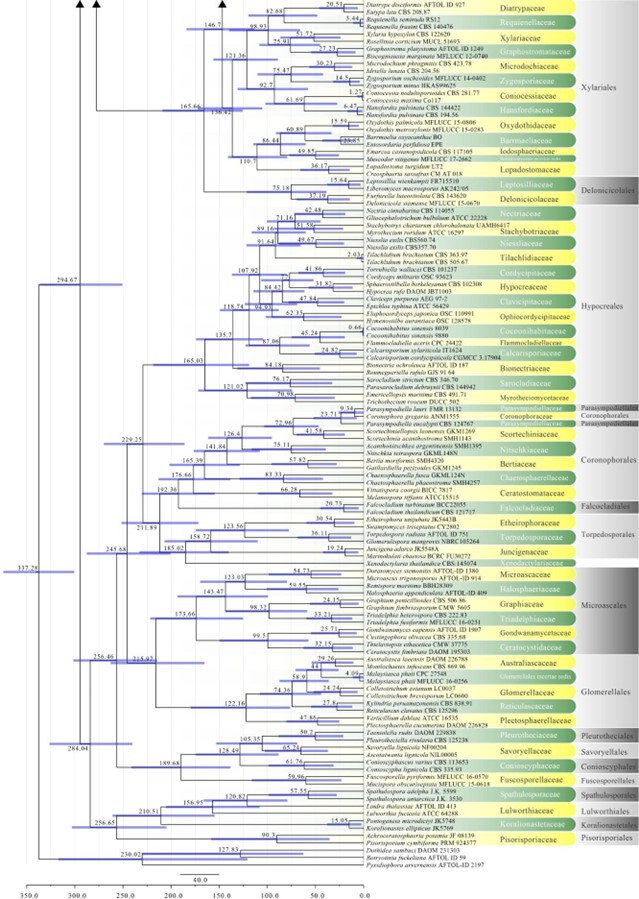
Figure 2 – Continued.
Families
Amphisphaeriales genus incertae sedis
