Allodiatrype arengae Konta & K.D. Hyde, sp. nov. Fig. 3
MycoBank number: MB 556929; Index Fungorum number: IF 556929; Facesoffungi number: FoF 05117;
Etymology: – Epithet refers to host genus, Arenga
Holotype: – MFLU 15-1444
Saprobic on petiole of Arenga pinnata (Arecaceae). Sexual morph: Stromata 690–940 μm long, 370–935 μm wide (x̄ = 830 × 700 μm, n = 10), with well-developed interior, solitary, superficial, black, without black stromatic, glabrous, subglobose to irregular, pustulate, 1–5- ascomata, with umbilicate ostioles appearing on the surface of the stroma. Ostiole opening through host bark and appearing as black spots, surrounded with a ring-like structure, composed of an outer layer of dark brown, small, tightly packed, thin parenchymatous cells and an inner layer of yellow, large, loosely packed, parenchymatous cells. Ascomata (excluding necks) 250–400 μm high, 240– 400 μm diam. (x̄ = 340 × 300 μm, n = 25), perithecial, immersed in the stroma, globose to subglobose, glabrous, ostioles individual, with a short neck. Ostiolar canal 100–170 μm high, 70– 130 μm diam. (x̄ = 130 × 100 μm, n = 20), cylindrical, sulcate, with periphyses. Peridium 12–25 μm wide, (x̄ = 20 μm, n = 40), composed of two sections, outer layer of brown to dark brown, thin- walled cells, arranged in textura angularis, inner layer of hyaline thin-walled cells of textura angularis. Hamathecium composed of 3–7 μm wide (x̄ = 5 μm, n = 40), septate, hyaline paraphyses. Asci (excluding stalks), spore-bearing part (14–)20(−45) × (4–)6–10(−12) μm (x̄ = 30 × 8 μm, n = 80), apically rounded, with J-apical ring, apex-bearing part (1.5–)3–5(−7.5) μm long (x̄ = 4 μm, n = 40), 8-spored, unitunicate, clavate, with long stalks, (28–)34–89(–103) μm long (x̄ = 64 μm, n = 60). Ascospores (6–)7–10(−12) × 2–3 μm (x̄ = 10 × 2 μm, n = 120), overlapping, yellowish to light-brown, ellipsoidal to cylindrical or elongate-allantoid, aseptate, smooth-walled. Asexual morph: Undetermined.
Geographical distribution: – Thailand.
Culture characters: – Ascospores germinated on MEA within 24 hours. Colonies on MEA, white in beginning, dense but thinning towards the edge, margin diffuse, reverse pale yellow in the middle (Fig. 3q).
Additional sequence data: – LSU (MN308402), SSU (MN308420), tef1 (MN525596), RPB2 (MN542886) (MFLUCC 15-0713).
Material examined: – THAILAND, Phang-Nga Province, on dead petiole of Arenga pinnata (Wurmb) Merr. (Arecaceae), 4 December 2014, S. Konta, PHR01b (MFLU 15-1444, holotype); ex- type living culture = MFLUCC 15-0713.
Notes: – Allodiatrype arengae was collected from a dead petiole of Arenga pinnata from Phang-Nga Province, Thailand. Allodiatrype arengae is phylogenetically distinct from its sister species A. elaeidicola, A. elaeidis, and A. thailandica with high statistical support (100% ML, 1.00 BYPP) (Fig. 1). A comparison of the nucleotide between Allodiatrype species and Diatrype disciformis is given in Table 6. Morphologically, A. arengae has superficial stromata lacking a black stromatic zone (Fig. 3a), while A. elaeidicola and A. elaeidis formed erumpent stromata, arising through the cracks in bark or epidermis with a black stromatic zone (Figs 4a, b, 5a), sometimes it covers the host surface (Fig. 4a, b). Other characters such as ascomata, asci and ascospores are mostly similar to A. thailandica, and their sizes also overlap. Our new strain is also recorded on a different host substrate from other species.
Table 6 Comparison of the nucleotides of Allodiatrype arengae to all species of Allodiatrype, and type species of the genus Diatrype.
| Species | LSU | SSU | ITS | TUB2 | RBP2 | tef1 | References |
| A. elaeidicola MFLUCC 15-0737 | 0 | 1/1030 (0.09%) | 13/605 (2.14%) | 28/1586 (1.76%) | 3/1139 (0.26%) | 0 | This study |
| A. elaeidis MFLUCC 15-0708 | 0 | 0 | 13/621 (2.09%) | 16/1579 (1.01%) | 2/1138 (0.17%) | 0 | This study |
| A. thailandica MFLUCC 14-1210 | – | – | 9/526 (1.71%) | – | – | – | Li et al. (2016) |
| A. thailandica MFLUCC 15-0711 | 0 | 9/1040 (0.86%) | 11/619 (1.77%) | – | – | – | This study |
| Diatrype disciformis T | 21/905 (2.32%) | 2/894 (0.22%) | 56/536 (10.44%) | – | 314/1137 (27.6%) | 69/957 (7.2%) | Acero et al. (2004) |
Notes – ‘-’ do not have sequence; ‘0’ no base pair similarity; T type species of Diatrype; base pair differences included gaps.
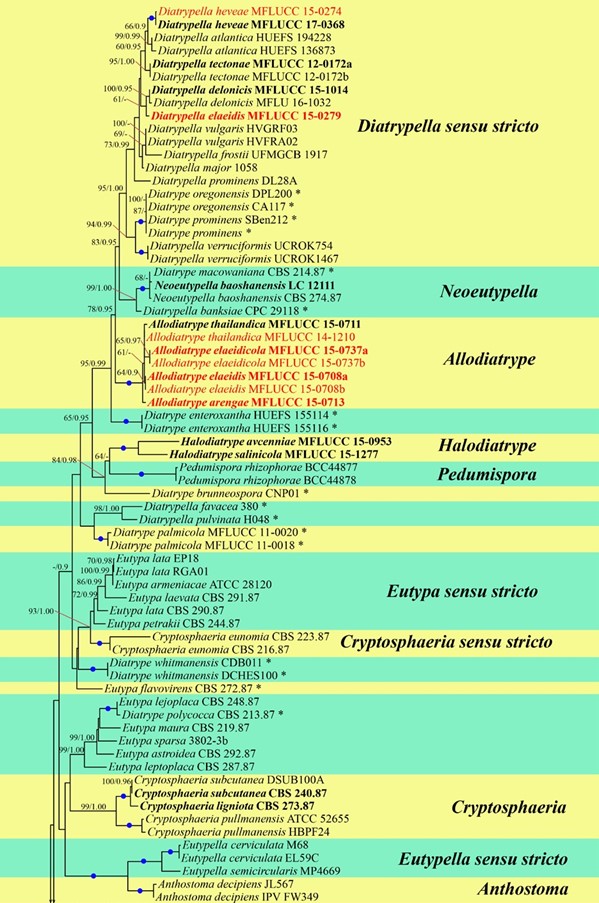
Figure 1 – Bayesian analyses the majority rule consensus tree of selected species in Diatrypaceae generated from combined ITS and TUB2 sequence data. Bootstrap support values for maximum likelihood (ML) greater than 50%, and Bayesian posterior probabilities (BYPP) greater than 0.90 are given at the nodes. Branches with 100% ML and 1.00 BYPP are shown with a blue dot. Ex-type strains are in bold. Newly generated sequences are in red. Novel taxa are in red bold. The asterisks represent unstable species.
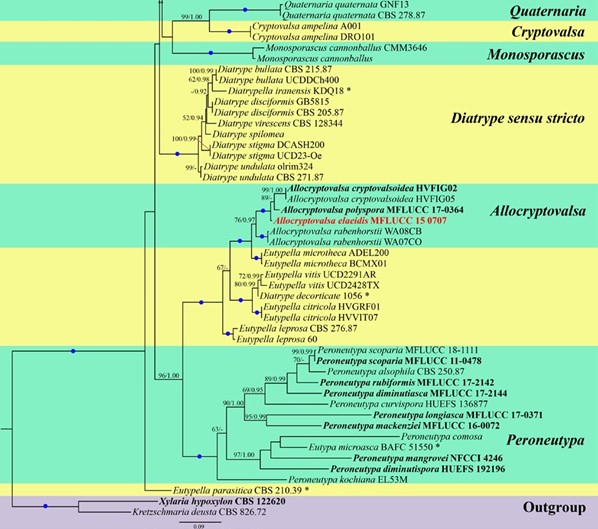
Figure 1 – Continued.
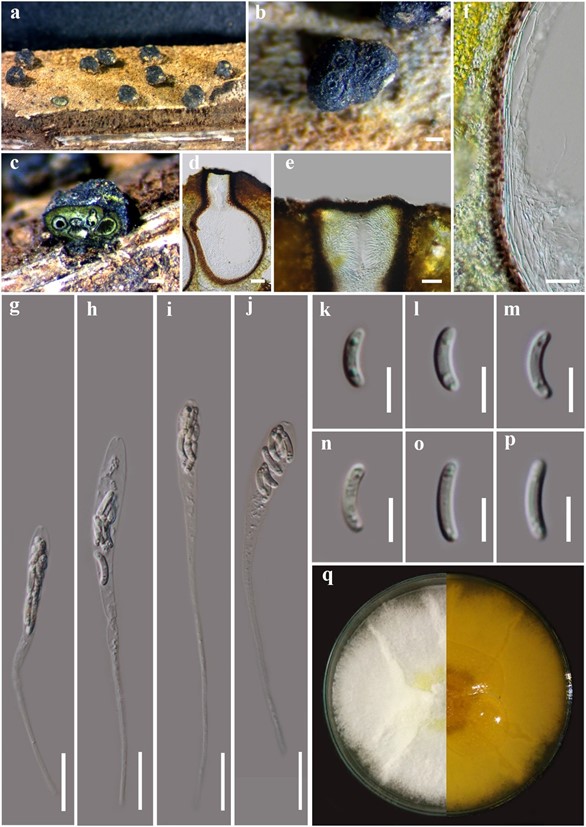
Figure 3 – Allodiatrype arengae (MFLU 15-1444, holotype) a Stromata on host substrate. b Close up of stroma (ostiole opening surrounded with a ring-like structure). c, d Section of stroma. e Ostiolar canal. f Peridium. g–j Asci. k–p Ascospores. q Colony on MEA. Bars: a = 500 μm, b, c = 200 μm, d, g–j = 50 μm, e, f = 20 μm, k–p = 5 μm.

Figure 4 – Allodiatrype elaeidicola (MFLU 15-1468, holotype) a Stromata on host substrate. b Close up of stromata (ostiole opening surrounded with a ring-like structure). c, d Section of stroma. e, i Mature asci. f–h Immature asci stained in cotton blue. j Ostiolar canal. k Peridium. l–q Ascospores. r Colony on MEA. Bars: a, b = 1000 μm, c = 500 μm, d = 200 μm, e–k = 50 μm, l–q = 5 μm.
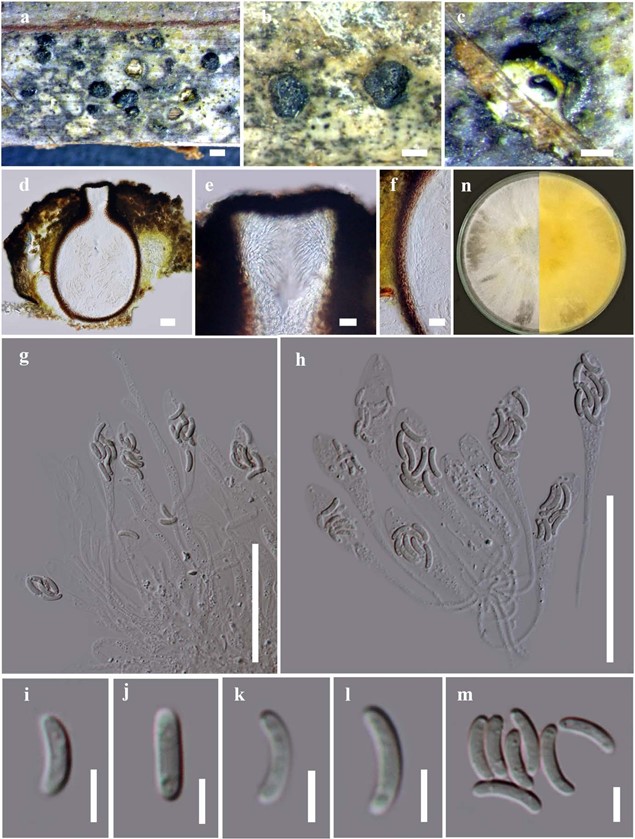
Figure 5 – Allodiatrype elaeidis (MFLU 15-1439, holotype) a Stromata on host substrate. b Close up of stromata (ostiole opening surrounded with a ring-like structure). c, d Section of stromata. e Ostiolar canal. f Peridium. g, h Asci. i–m Ascospores. n Colony on MEA. Bars: a, b = 500 μm, c = 200 μm, d, g, h = 50 μm, e, f = 20 μm, i–m = 5 μm.
