Beltraniaceae Nann., Repert. mic. uomo: 498 (1934)
MycoBank number: MB 80516; Index Fungorum number: IF 80516; Facesoffungi number: FoF 05312; 80 species.
Saprobic on plant tissues. Sexual morph: Ascomata pale yellow, solitary to aggregated on OA and PDA, globose to somewhat papillate, with central ostiole; wall of 3–4 layers of subhyaline cells of textura angularis to intricata. Paraphyses septate, cellular, anastomosing, distributed among asci. Asci 8-spored, unitunicate, subcylindrical, sessile. Ascospores tri- to multi-seriate, hyaline, obovoid, granular, smooth, aseptate with dissolving mucoid sheath. Asexual morph: Hyphomycetous. Mycelium immersed to superficial, composed of subhyaline to brown, thin-walled hyphae. Stromata usually present, parenchymatous to pseudoparenchymatous, hyaline to brown, often confined to epidermal cells. Setae present or absent, straight, thick-walled, dark brown, smooth or verrucose, with radially lobed basal cell, tapering to acute apex. Conidiophores simple or sometimes branched, erect, septate, pale brown, arising from the base of setae or separate, with or without radially lobed basal cell. Conidiogenous cells holoblastic, polyblastic, pale brown, integrated, denticulate. Conidia biconic, lageniform to navicular, subhyaline to red-brown, with transverse band of pale pigment at widest part of the conidium, base rounded, 1-denticulate or rostrate, apex spicate, apiculate or truncate (adapted from Crous et al. (2015a) and Maharachchikumbura et al. (2016b)).
Type genus – Beltrania Penz.
Notes – Beltrania, Beltraniella, Beltraniopsis, Hemibeltrania, Parapleurotheciopsis, Porobeltraniella, Pseudobeltrania, Pseudosubramaniomyces and Subsessila, are accepted in the family (Maharachchikumbura et al. 2015, 2016b, Hongsanan et al. 2017, Lin et al. 2017a). We introduce and illustrate a new species, Beltraniella ramosiphora based on phylogenetic data (Figs. 4, 45).
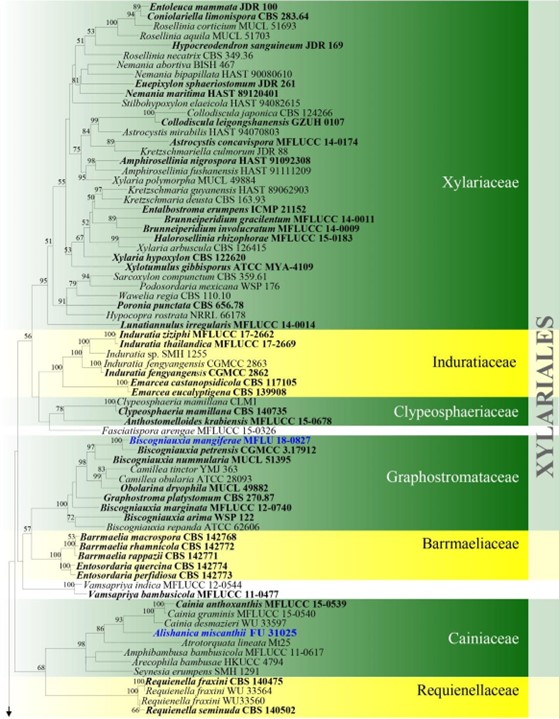
Figure 4 – Phylogram generated from maximum likelihood analysis based on combined ITS, LSU, rpb2 and tub2 sequence data for Xylariomycetidae. Two hundred and seventy-two strains are included in the combined analyses which comprised 4211 characters (1168 characters for ITS, 937 characters for LSU, 1128 characters for rpb2, 978 characters for tub2) after alignment. Achaetomium macrosporum (CBS 532.94), Chaetomium elatum (CBS 374.66) and Sordaria fimicola (CBS 723.96) are outgroup taxa. Single gene analyses were carried out and the topology of each tree had clade stability. The best RaxML tree with a final likelihood value of – 132297.706952 is presented. Estimated base frequencies were as follows: A = 0.241914, C = 0.251908, G = 0.265558, T = 0.240620; substitution rates AC = 1.281946, AG = 3.512297, AT = 1.499895, CG = 1.121065, CT = 6.472834, GT = 1.000000; gamma distribution shape parameter a = 0.678614. Bootstrap support values for ML greater than 75% are given near the nodes. Ex-type strains are in bold. The newly generated sequences are indicated in blue.
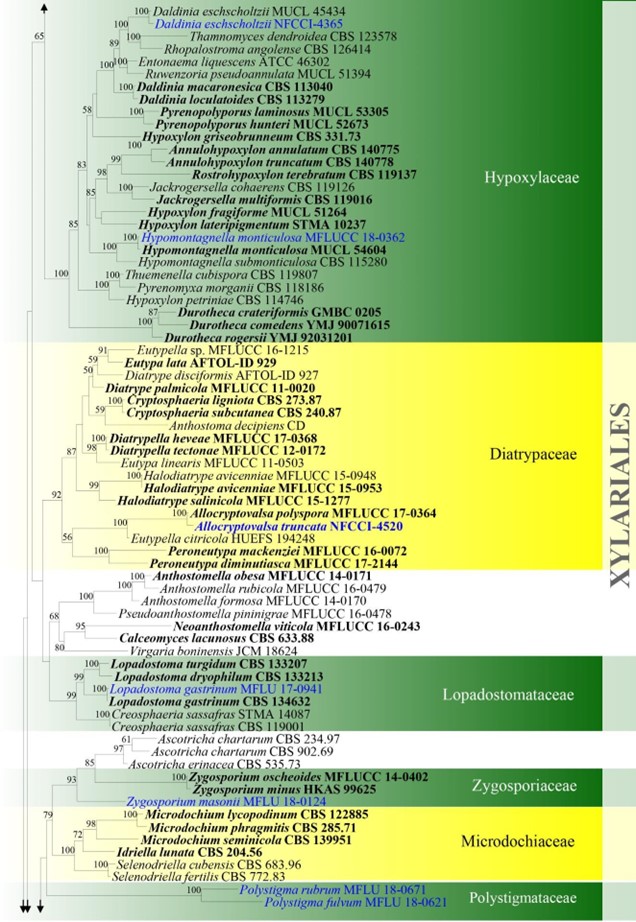
Figure –4 Continued.
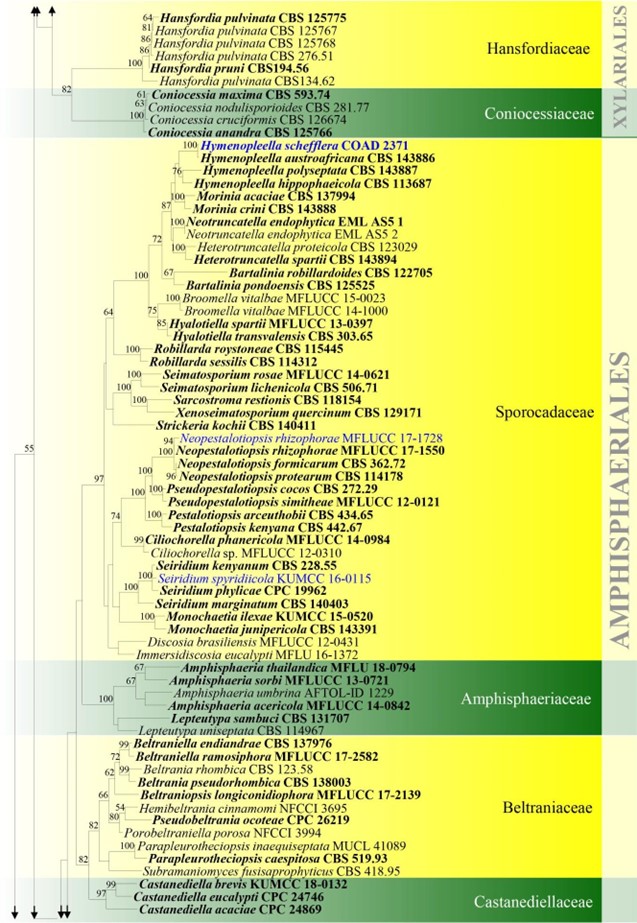
Figure –4 Continued.

Figure –4 Continued.
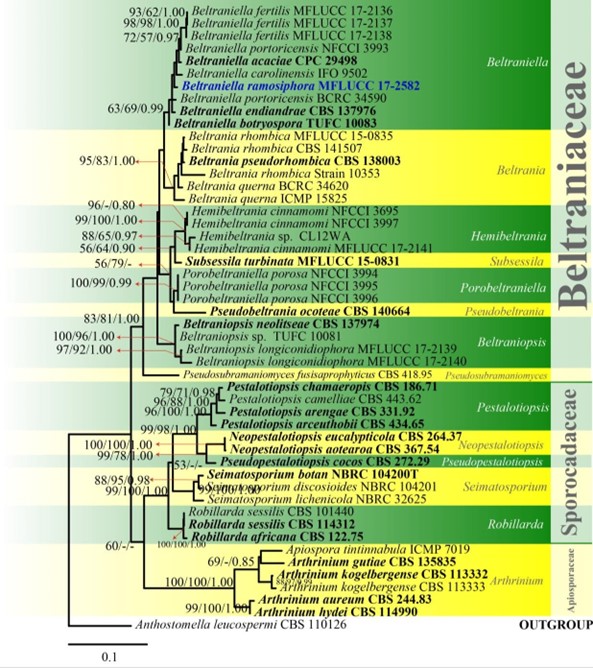
Figure 45 – Phylogram generated from maximum likelihood analysis based on combined LSU and ITS sequence data of Beltraniaceae. Fifty strains are included in the combined analyses which comprised 1546 characters (884 characters for LSU, 662 characters for ITS) after alignment. Tree topology of the maximum likelihood analysis is similar to the Bayesian and maximum parsimony analyses. The best RaxML tree with a final likelihood value of -7145.370801 is presented. Estimated base frequencies were as follows: A = 0.252192, C = 0.214861, G = 0.266786, T = 0.266162; substitution rates AC = 1.363640, AG = 2.557848, AT = 1.973142, CG = 1.035845, CT = 5.942470, GT = 1.000000; gamma distribution shape parameter α = 0.171878. Bootstrap support values for maximum likelihood and maximum parsimony greater than 50% and Bayesian posterior probabilities greater than 0.8 are indicated above or below the nodes as MLBS/MPBS/PP. The tree is rooted with Anthostomella leucospermi (CBS 110126). Ex-type strains are in bold and black. The newly generated sequences are indicated in blue.
